Gather & Spread
Overview
Teaching: 30 min
Exercises: 15 minQuestions
How can I change the format of dataframes?
Objectives
To understand the concepts of ‘long’ and ‘wide’ data formats and be able to convert between them with tidyr.
Having had some exposure to working with tidyverse functions, you might now understand why it is beneficial to work with tidy data. These data manipulation functions are all designed to work easily with tidy data, leaving you with more time to actually try to answer questions with the data.
Unfortunately, real data sets can come in all shapes and sizes and may not be structured for you to
efficiently analyse it. To help tidy our data up in preparation for analysis, we will turn to the
tidyr package from the tidyverse.
Tidy data review
Before getting into a real example, we’ll have a look at a toy dataset. The following dataframe has two weight measurements for three different cows. Have a look at the way the data is structured. Is it tidy?
cows <- data_frame(id = c(1, 2, 3),
weight1 = c(203, 227, 193),
weight2 = c(365, 344, 329))
cows
# A tibble: 3 × 3
id weight1 weight2
<dbl> <dbl> <dbl>
1 1 203 365
2 2 227 344
3 3 193 329
What might make this dataset tidy?
Table massaging with gather() and spread()
To make this dataframe tidy, we may need to have only one weight variable, and another variable
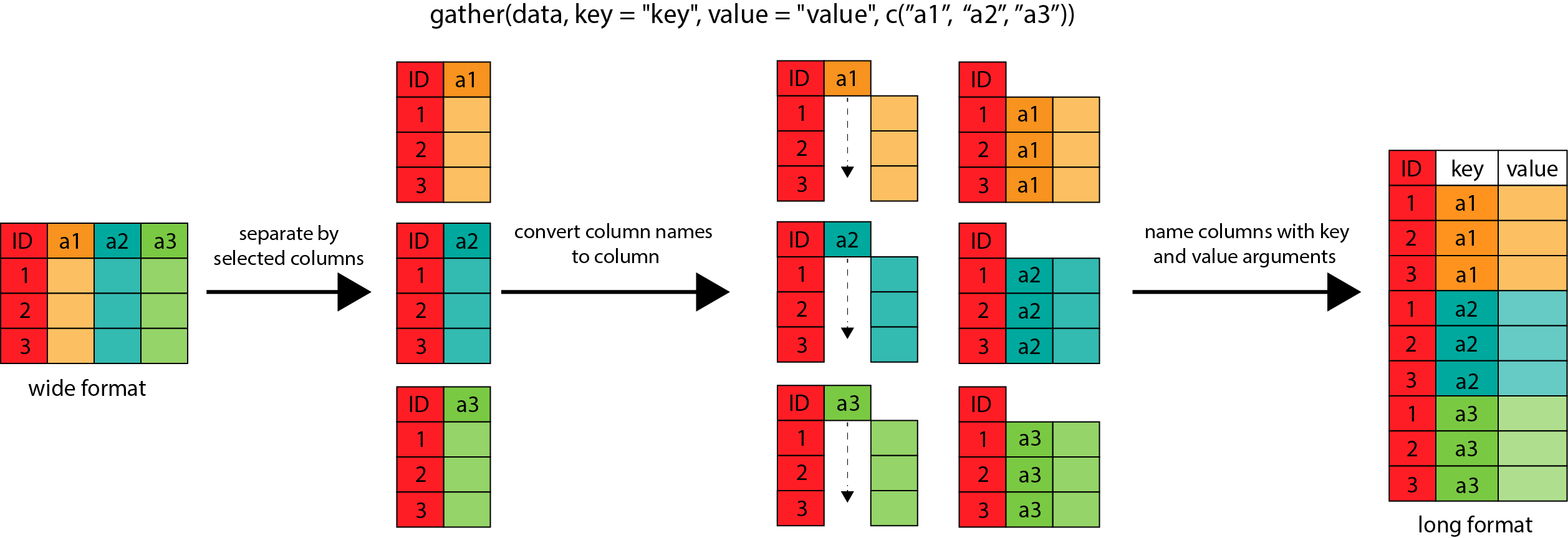
describing whether it is measurement 1 or 2. The gather() function from tidyr can do this
conversion for us. You can think of gather() as scooping all the data up into one big tall pile.
cows_long <- cows %>%
gather(measurement, weight, weight1, weight2)
cows_long
# A tibble: 6 × 3
id measurement weight
<dbl> <chr> <dbl>
1 1 weight1 203
2 2 weight1 227
3 3 weight1 193
4 1 weight2 365
5 2 weight2 344
6 3 weight2 329
gather() works by converting the data into a set of key-value pairs, in which the
key describes what the data is, and the value records the actual data. For example, weight1-203
is the key-value pair for the first row of the gathered table.
The call to gather() therefore works in two parts. First we specify the names of the new key-value pair
columns (measurement and weight in this case). Then we tell it which columns should be gathered
(weight1 and weight2 here). gather() then takes the contents of those columns and places them
into the new value column, and labels them with their column name as the key.
This transformation may be easier to see in action, so let’s have a look at the way gather() has
worked on the original data.
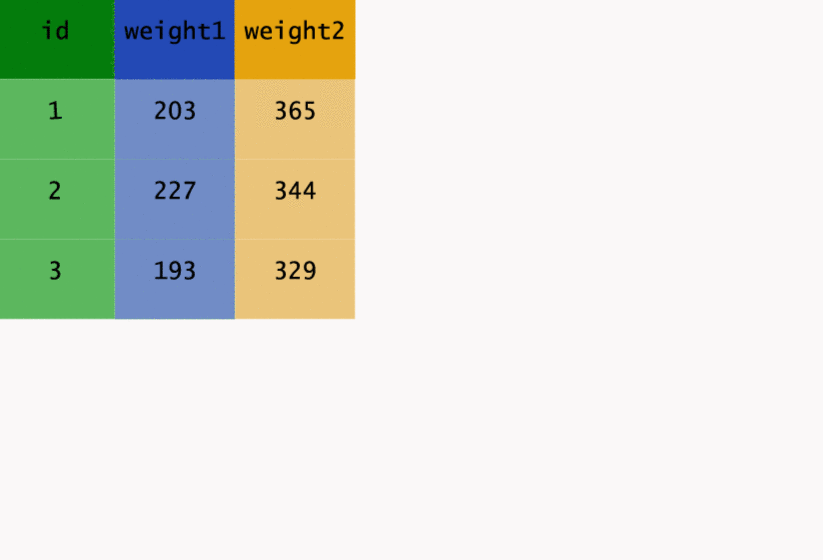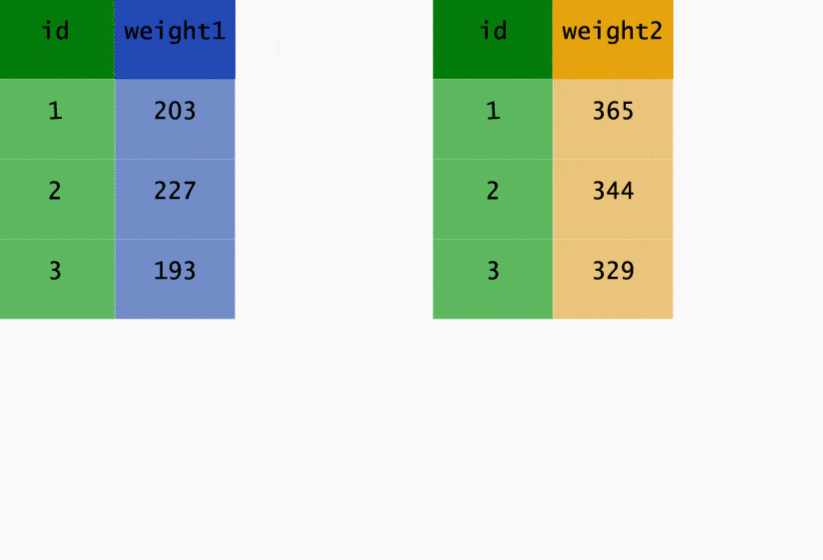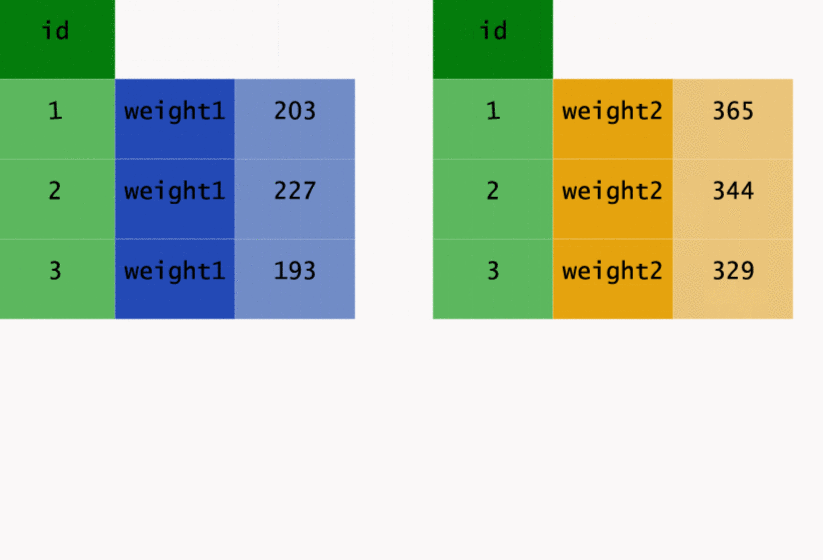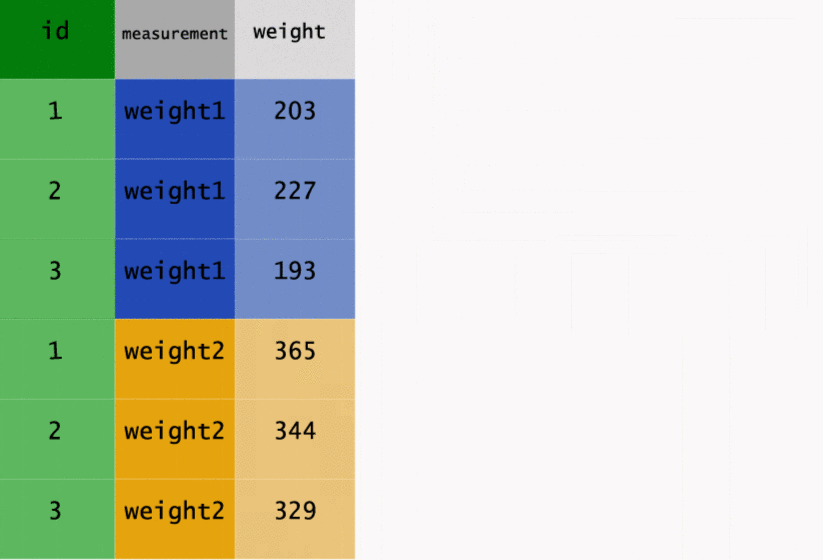
As well as going from wide to long, we can go back the other way. Instead of gather(), we need to
spread() the data.
cows_long %>%
spread(measurement, weight)
# A tibble: 3 × 3
id weight1 weight2
<dbl> <dbl> <dbl>
1 1 203 365
2 2 227 344
3 3 193 329
This works in reverse, taking the names of out key-value columns and spreading them out. Each unique value in the key column is given a separate column of it’s own. And the content of that column is taken from the specified value column.
Realistic data
Until now, we’ve been using the original gapminder dataset which comes pre-tidied, but ‘real’ data (i.e. our own research data) will never be so well organised. Here let’s start with the wide format version of the gapminder dataset.
Download the wide version of the gapminder data from here and save it in your project’s
datadirectory.
Read in the wide gapminder and have a look at it.
gap_wide <- read_csv("data/gapminder_wide.csv")
gap_wide
# A tibble: 142 × 38
continent country gdpPercap_1952 gdpPercap_1957 gdpPercap_1962 gdpPercap_1967
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Africa Algeria 2449. 3014. 2551. 3247.
2 Africa Angola 3521. 3828. 4269. 5523.
3 Africa Benin 1063. 960. 949. 1036.
4 Africa Botswa… 851. 918. 984. 1215.
5 Africa Burkin… 543. 617. 723. 795.
6 Africa Burundi 339. 380. 355. 413.
7 Africa Camero… 1173. 1313. 1400. 1508.
8 Africa Centra… 1071. 1191. 1193. 1136.
9 Africa Chad 1179. 1308. 1390. 1197.
10 Africa Comoros 1103. 1211. 1407. 1876.
# ℹ 132 more rows
# ℹ 32 more variables: gdpPercap_1972 <dbl>, gdpPercap_1977 <dbl>,
# gdpPercap_1982 <dbl>, gdpPercap_1987 <dbl>, gdpPercap_1992 <dbl>,
# gdpPercap_1997 <dbl>, gdpPercap_2002 <dbl>, gdpPercap_2007 <dbl>,
# lifeExp_1952 <dbl>, lifeExp_1957 <dbl>, lifeExp_1962 <dbl>,
# lifeExp_1967 <dbl>, lifeExp_1972 <dbl>, lifeExp_1977 <dbl>,
# lifeExp_1982 <dbl>, lifeExp_1987 <dbl>, lifeExp_1992 <dbl>, …
You can see that wide format has many more columns than our original gapminder data because each metric has a separate column for each year of measurement.
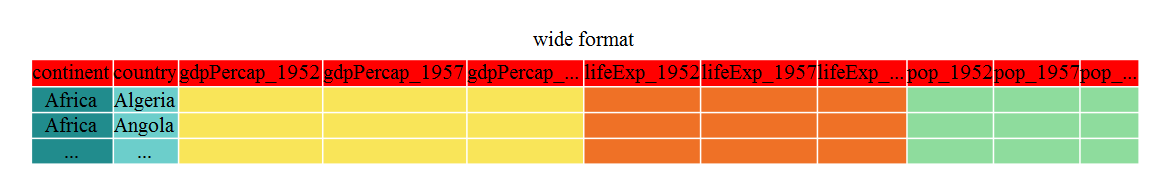
From wide to long
The first step towards recreating the nice and tidy gapminder format is to
convert this data from a wide to a long format. The tidyr function gather() will
‘gather’ your observation variables into a single variable.
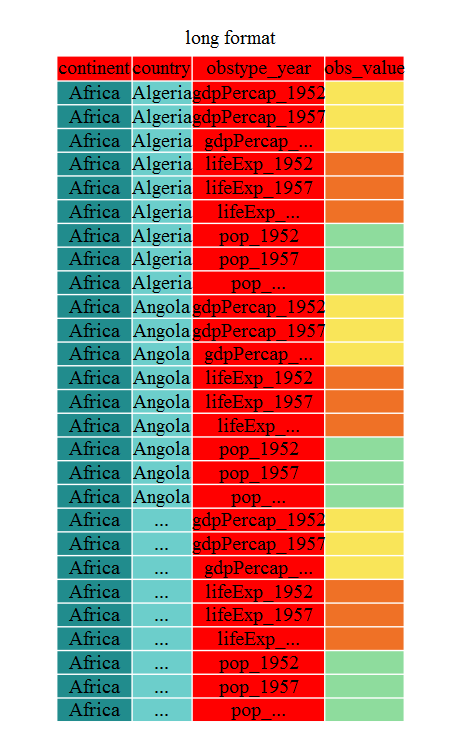
gap_long <- gap_wide %>%
gather(obstype_year, obs_values, starts_with('pop'),
starts_with('lifeExp'), starts_with('gdpPercap'))
gap_long
# A tibble: 5,112 × 4
continent country obstype_year obs_values
<chr> <chr> <chr> <dbl>
1 Africa Algeria pop_1952 9279525
2 Africa Angola pop_1952 4232095
3 Africa Benin pop_1952 1738315
4 Africa Botswana pop_1952 442308
5 Africa Burkina Faso pop_1952 4469979
6 Africa Burundi pop_1952 2445618
7 Africa Cameroon pop_1952 5009067
8 Africa Central African Republic pop_1952 1291695
9 Africa Chad pop_1952 2682462
10 Africa Comoros pop_1952 153936
# ℹ 5,102 more rows
Inside gather() we first name the new column for the new ID variable (obstype_year), the name
for the new amalgamated observation variable (obs_value), then the names of the old observation
variable. We could have typed out all the observation variables, but since gather() can use all the same
helper functions
as the select() function can, we can use the starts_with() argument to select all variables
that starts with the desired character string. Gather also allows the alternative
syntax of using the - symbol to identify which variables are not to be
gathered (i.e. ID variables) so that
gap_long <- gap_wide %>%
gather(obstype_year, obs_values, -continent, -country)
is an alternative way of specifying the columns to gather in the wide data set. The best method for any particular data set will depend on how many columns you need to gather and how they are named.
Separating column data
Now, the obstype_year column actually contains two pieces of information, the observation
type (pop,lifeExp, or gdpPercap) and the year. We can use the separate() function to split
the character strings into multiple variables.
gap_separated <- gap_long %>%
separate(obstype_year, into = c('obs_type', 'year'), sep = "_")
gap_separated
# A tibble: 5,112 × 5
continent country obs_type year obs_values
<chr> <chr> <chr> <chr> <dbl>
1 Africa Algeria pop 1952 9279525
2 Africa Angola pop 1952 4232095
3 Africa Benin pop 1952 1738315
4 Africa Botswana pop 1952 442308
5 Africa Burkina Faso pop 1952 4469979
6 Africa Burundi pop 1952 2445618
7 Africa Cameroon pop 1952 5009067
8 Africa Central African Republic pop 1952 1291695
9 Africa Chad pop 1952 2682462
10 Africa Comoros pop 1952 153936
# ℹ 5,102 more rows
You provide separate() with a column name containing the values to split, the names of the columns
you would like to separate them into, and where to split the values (by default it splits on any non
alphanumeric character).
Challenge 1
If you look at the table above, you will see that
separate()creates character columns out of both theobs_typeandyearcolumns, but our original gapminder has year as a numeric. How would you fix this discrepancy?Hint: Read through the arguments for
separatewith?separateSolution to Challenge 1
The
convertargument will try to convert character strings to other data types when set toTRUEgap_separated <- gap_long %>% separate(obstype_year, into = c('obs_type', 'year'), sep = "_", convert = TRUE) gap_separated# A tibble: 5,112 × 5 continent country obs_type year obs_values <chr> <chr> <chr> <int> <dbl> 1 Africa Algeria pop 1952 9279525 2 Africa Angola pop 1952 4232095 3 Africa Benin pop 1952 1738315 4 Africa Botswana pop 1952 442308 5 Africa Burkina Faso pop 1952 4469979 6 Africa Burundi pop 1952 2445618 7 Africa Cameroon pop 1952 5009067 8 Africa Central African Republic pop 1952 1291695 9 Africa Chad pop 1952 2682462 10 Africa Comoros pop 1952 153936 # ℹ 5,102 more rows
The opposite to separate() is unite(). Use this when you are wanting to combine columns into
one long string. You provide unite() with the name to give the newly combined column, and the
names of the columns to combine.
gap_separated %>%
unite(obstype_year, obs_type, year)
# A tibble: 5,112 × 4
continent country obstype_year obs_values
<chr> <chr> <chr> <dbl>
1 Africa Algeria pop_1952 9279525
2 Africa Angola pop_1952 4232095
3 Africa Benin pop_1952 1738315
4 Africa Botswana pop_1952 442308
5 Africa Burkina Faso pop_1952 4469979
6 Africa Burundi pop_1952 2445618
7 Africa Cameroon pop_1952 5009067
8 Africa Central African Republic pop_1952 1291695
9 Africa Chad pop_1952 2682462
10 Africa Comoros pop_1952 153936
# ℹ 5,102 more rows
From long to tidy
The final step in recreating the gapminder data structure is to spread our observation variables out from the long format we have created.
Challenge 2
Spread the
gap_separateddata above to create a new data frame that has the same dimensions as the originalgapminderdata.Solution to Challenge 2
gap_orig <- gap_separated %>% spread(obs_type, obs_values) gap_orig# A tibble: 1,704 × 6 continent country year gdpPercap lifeExp pop <chr> <chr> <int> <dbl> <dbl> <dbl> 1 Africa Algeria 1952 2449. 43.1 9279525 2 Africa Algeria 1957 3014. 45.7 10270856 3 Africa Algeria 1962 2551. 48.3 11000948 4 Africa Algeria 1967 3247. 51.4 12760499 5 Africa Algeria 1972 4183. 54.5 14760787 6 Africa Algeria 1977 4910. 58.0 17152804 7 Africa Algeria 1982 5745. 61.4 20033753 8 Africa Algeria 1987 5681. 65.8 23254956 9 Africa Algeria 1992 5023. 67.7 26298373 10 Africa Algeria 1997 4797. 69.2 29072015 # ℹ 1,694 more rowsdim(gap_orig)[1] 1704 6dim(gapminder)[1] 1704 6
We’re almost there, the original was sorted by country, continent, then year. So finish
everything off with an arrange() and then a select() to get the columns in the right order.
gap_orig <- gap_orig %>%
arrange(country, continent, year) %>%
select(country, continent, year, lifeExp, pop, gdpPercap)
gap_orig
# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<chr> <chr> <int> <dbl> <dbl> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779.
2 Afghanistan Asia 1957 30.3 9240934 821.
3 Afghanistan Asia 1962 32.0 10267083 853.
4 Afghanistan Asia 1967 34.0 11537966 836.
5 Afghanistan Asia 1972 36.1 13079460 740.
6 Afghanistan Asia 1977 38.4 14880372 786.
7 Afghanistan Asia 1982 39.9 12881816 978.
8 Afghanistan Asia 1987 40.8 13867957 852.
9 Afghanistan Asia 1992 41.7 16317921 649.
10 Afghanistan Asia 1997 41.8 22227415 635.
# ℹ 1,694 more rows
gapminder
# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779.
2 Afghanistan Asia 1957 30.3 9240934 821.
3 Afghanistan Asia 1962 32.0 10267083 853.
4 Afghanistan Asia 1967 34.0 11537966 836.
5 Afghanistan Asia 1972 36.1 13079460 740.
6 Afghanistan Asia 1977 38.4 14880372 786.
7 Afghanistan Asia 1982 39.9 12881816 978.
8 Afghanistan Asia 1987 40.8 13867957 852.
9 Afghanistan Asia 1992 41.7 16317921 649.
10 Afghanistan Asia 1997 41.8 22227415 635.
# ℹ 1,694 more rows
Great work! We’ve taken a messy data set and tidied it into a form that works well with all the tidyverse functions you have been using up until now.
Challenge 3
Practice your
gathering skills with the inbuilttidyr::table4adata frame, which contains the number of TB cases recorded by the WHO in three countries in 1999 and 2000. Is the data frame untidy, and can you tidy it withgather()?tidyr::table4a# A tibble: 3 × 3 country `1999` `2000` <chr> <dbl> <dbl> 1 Afghanistan 745 2666 2 Brazil 37737 80488 3 China 212258 213766Solution to Challenge 3
gather(tidyr::table4a, key = year, value = TB_cases, -country)# A tibble: 6 × 3 country year TB_cases <chr> <chr> <dbl> 1 Afghanistan 1999 745 2 Brazil 1999 37737 3 China 1999 212258 4 Afghanistan 2000 2666 5 Brazil 2000 80488 6 China 2000 213766
Challenge 4
Practice your
spreading skills with the inbuilttidyr::table2data frame, which contains the number of TB cases recorded by the WHO in three countries in 1999 and 2000 as well as their population. Is the data frame untidy, and can you tidy it withspread()?tidyr::table2# A tibble: 12 × 4 country year type count <chr> <dbl> <chr> <dbl> 1 Afghanistan 1999 cases 745 2 Afghanistan 1999 population 19987071 3 Afghanistan 2000 cases 2666 4 Afghanistan 2000 population 20595360 5 Brazil 1999 cases 37737 6 Brazil 1999 population 172006362 7 Brazil 2000 cases 80488 8 Brazil 2000 population 174504898 9 China 1999 cases 212258 10 China 1999 population 1272915272 11 China 2000 cases 213766 12 China 2000 population 1280428583Solution to Challenge 4
spread(tidyr::table2, key = type, value = count)# A tibble: 6 × 4 country year cases population <chr> <dbl> <dbl> <dbl> 1 Afghanistan 1999 745 19987071 2 Afghanistan 2000 2666 20595360 3 Brazil 1999 37737 172006362 4 Brazil 2000 80488 174504898 5 China 1999 212258 1272915272 6 China 2000 213766 1280428583
Key Points
Use the tidyr package to change the layout of dataframes.
Use
gather()to go from wide to long format.Use
spread()to go from long to wide format.