Content from Introduction
Last updated on 2024-01-22 | Edit this page
Estimated time: 15 minutes
Overview
Questions
- How can we use computers more effectively in a scientific project?
Objectives
- Discuss what can go wrong and right in scientific computing
This lesson covers a set of good computing practices that every researcher can adopt, regardless of computational skill. Informally, it teaches how to not lose your stuff, and generally how to be more efficient.
Many of our recommendations are for the benefit of the collaborator every researcher cares about most: their future self (as the joke goes, yourself from 3 months ago doesn’t answer email…).
Practically, future you will either:
- curse current you (bad), or
- thank current you (better).
Adopting good practices is kind to your future self.
So, which habits and practices can save you time in the future and improve your work?
Discuss in groups
- What can go wrong in research computing?
- What can go right in research computing?
Consider the entire data life cycle of a project, as well as the actual analysis.
- Planning and designing
- Collecting and capturing
- Organizing and storing
- Interpreting and analyzing
- Managing and preserving
- Publishing and sharing
- You can lose your data
- You can re-analyze your data in 1 year’s time and learn something important
- And many, many, more examples
Challenges in data-heavy biology
In biology, as in other sciences:
- All your raw data are digital files
- How do you manage them?
- There are many tools to process data
- Too much choice, many bad choices
- e.g. Excel changes gene names, and loses COVID test results…
- There are many levels at which to study a problem and many steps to
understand
- Where do you even start?
Good news: everyone has these problems!
- Other people have thought about good practices and created good tools.
- You don’t have to reinvent practices and tools.
- You can learn to be “good enough” in scientific computing.
- This is an ongoing process through your career.
Principles: planning, modular organization, names, documentation
This lesson has episodes covering data management, software, project organization, collaboration, keeping track of changes, and manuscripts.
Good Enough Practices rely on a shared set of principles that span these areas:
- Planning: plan out how to work. Any plan that you can stick to is better than no plan.
- Modular organization: organize your data, code, and projects into coherent modules.
- Names: give good names to your files, folders, and functions, that make them easy to find and to understand.
- Documentation: explicitly write down everything you and your collaborators need to know in the future.
Key Points
- Computing is essential in science and (almost) all data are digital
- A set of good enough practices can make you more efficient
- Future you will thank past you for adopting good practices
- Shared Principles: planning, modular organisation, names, documentation
Content from Data Management
Last updated on 2024-01-28 | Edit this page
Estimated time: 32 minutes
Overview
Questions
- What is data management?
- What data should I back up, and how?
- How can I share my data effectively?
Objectives
- Identify problems with data management practices
- Understand what raw data is
- Understand what backing up data means and why it is important to back up in more than one location
- Be able to decide on appropriate file names and identifiers
- Be able to create analysis ready datasets
- Understand the importance of documenting your process
- Understand what a DOI is and its usefulness
Data management
Data within a project may need to exist in various forms, ranging from what first arrives to what is actually used for the primary analyses. Data management is the process of storing, documenting, organizing, and sharing the data created and collected during a project. Our recommendations have two main themes. One is to work towards ready-to-analyze data incrementally, documenting both the intermediate data and the process. We also describe the key features of “tidy data”, which can be a powerful accelerator for analysis [wickham2014, hart2016].
Data management problems (5 minutes)
In your opinion, what can go wrong when you have poor data management? Write down 2 issues in the collaborative document.
- Data loss
- Data corruption, making data unusable
- Running out of storage capacity, making it hard to save data
- Confusion: what does this data mean? where does it come from? what is its purpose?
- Versioning issues: Which version of the data made this figure? Which version of the analysis script was used for the manuscript?
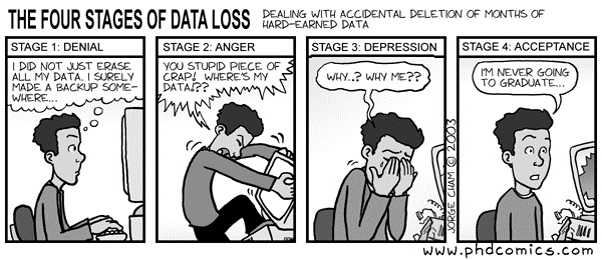
Save the raw data
Backing up your data (5 minutes)
Which of the following do you believe are good ways and bad ways of backing up your data?
- Commercial cloud service
- In-house cloud service (operates similarly to a commercial cloud service but with servers and infrastructure maintained by your organization)
- USB pen-drive
- External hard-drive
- My laptop
- My workstation’s hard-disk
- Network drive
- Commercial cloud service: it depends. Where are the servers located? How secure is it? How reliable is it? Do you have to pay for the service and what happens to your data if you can no longer afford it?
- In-house cloud service: this is a good way to back up your data (usually). You have local support. It is probably compliant with funders and data security guidelines for most data sets. If you work with particularly sensitive data (for example data of patients), we suggest still discussing with IT and/or data security officers.
- USB pen drive: definitely not! Pen-drives are prone to dying (and your data with it). It also raises data security issues and they can be easily lost.
- External hard-drive: see above.
- My laptop: it is good as a temporal storage solution for your active data. However, you should back it up appropriately.
- My workstation’s hard-disk: it is good as a temporal storage solution for your active data. However, you should back it up appropriately.
- Network drive: this is a good way to back up your data (usually). You have local support. It is probably compliant with funders and data security guidelines.
Backing up your data is essential, otherwise it is a question of when (not if) you lose it.
Where possible, save data as originally generated (i.e. by an instrument or from a survey). It is tempting to overwrite raw data files with cleaned-up versions, but faithful retention is essential for re-running analyses from start to finish; for recovery from analytical mishaps; and for experimenting without fear. Consider changing file permissions to read-only or using spreadsheet protection features, so it is harder to damage raw data by accident or to hand edit it in a moment of weakness.
Some data will be impractical to manage in this way. For example, you should avoid making local copies of large, stable databases. In that case, record the exact procedure used to obtain the raw data, as well as any other pertinent information, such as an official version number or the date of download.
In CSIRO, both raw data and final project data can (and should) be backed up on the Data Access Portal (it can host internal-only data copies as well as publicly published data). We also have project-allocated Bowen storage and HPC-linked high-volume storage paths that IM&T can provide use of, but check if any automated backup schedules are in place.
Also, remember not to place data in Git repositories. These are for code, scripts, notes, documentation, etc., but not data. Good data curation practices are different to good code curation practices.
Working with sensitive data
Identify whether your project will work with sensitive data - by which we might mean:
- Research data including personal data or identifiers (this might include names and addresses, or potentially identifyable genetic data or health information, or confidential information)
- Commercially sensitive data or information (this might include intellectual property, or data generated or used within a restrictive commercial research funding agreement)
- Data which may cause harm or adverse affects if released or made public (for example data relating to rare or endangered species which could cause poaching or fuel illegal trading)
It is important to understand the restrictions which may apply when working with sensitive data, and also ensure that your project complies with any applicable laws relating to storage, use and sharing of sensitive data (for example, laws like the General Data Protection Regulation, known as the GDPR). These laws vary between countries and may affect whether you can share information between collaborators in different countries.
You should have completed some compulsory online training on working
with sensitive data within CSIRO.
The Australian
Research Data Commons \(ARDC\)
provides some additional guides for working with sensitive data within
Australia.
Create the data you wish to see in the world
Filenames: Store especially useful metadata as part of the
filename itself, while keeping the filename regular enough for easy
pattern matching. For example, a filename like
2016-05-alaska-b.csv makes it easy for both people and
programs to select by year or by location. Common file naming
conventions are discussed in the Turing
Way and in the Project
Organization episode of this lesson.
Variable names: Replace inscrutable variable names and
artificial data codes with self-explaining alternatives, e.g., rename
variables called name1 and name2 to
first_name and family_name, recode the
treatment variable from 1 vs. 2 to
untreated vs. treated, and replace artificial
codes for missing data, such as “-99”, with NA, a code used
in most programming languages to indicate that data is “Not Available”
[white2013].
File formats: Convert data from closed, proprietary formats to open, non-proprietary formats that ensure machine readability across time and computing setups [UIllinois]. Good options include CSV for tabular data, JSON, YAML, or XML for non-tabular data such as graphs, and HDF5 for certain kinds of structured data.
Create the dataset you wish you had received. The goal here is to improve machine and human readability, but not to do vigorous data filtering or add external information. Machine readability allows automatic processing using computer programs, which is important when others want to reuse your data. Specific examples of non-destructive transformations that we recommend at the beginning of analysis:
- Create analysis-friendly data
- Record all the steps used to process data
- Record different data types in individual tables as appropriate (e.g. sample metadata may be kept separately from sequencing experiment metadata)
- Use unique identifiers for every record in a table, allowing linkages between tables (e.g. sample identifiers are recorded in the sequencing experiment metadata)
Create analysis-friendly data
Discussion (2 minutes)
Which of the table layouts is analysis friendly? Discuss. 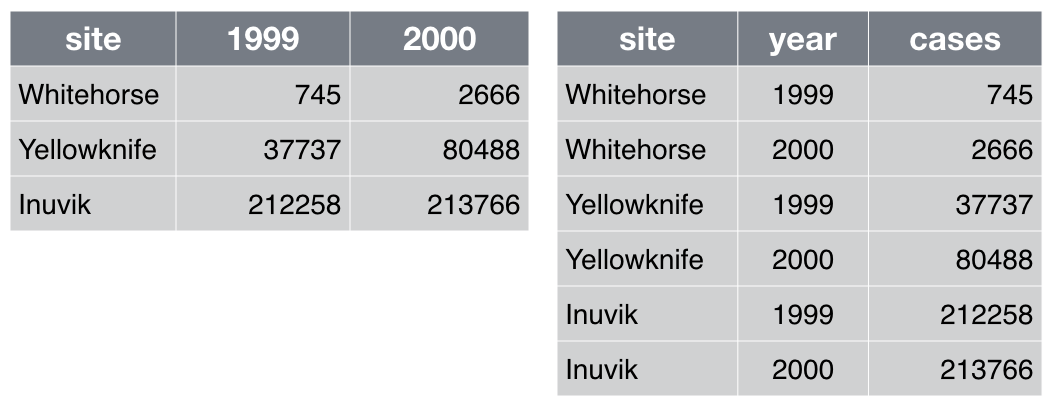
Analysis can be much easier if you are working with so-called “tidy” data [wickham2014]. Two key principles are:
Make each column a variable: Don’t cram two variables into one, e.g., “male_treated” should be split into separate variables for sex and treatment status. Store units in their own variable or in metadata, e.g., “3.4” instead of “3.4kg”.
Make each row an observation: Data often comes in a wide format, because that facilitated data entry or human inspection. Imagine one row per field site and then columns for measurements made at each of several time points. Be prepared to gather such columns into a variable of measurements, plus a new variable for time point. The figure above presents an example of such a transformation.
Record all the steps used to process data
Data manipulation is as integral to your analysis as statistical modeling and inference. If you do not document this step thoroughly, it is impossible for you, or anyone else, to repeat the analysis.
The best way to do this is to write scripts for every stage of data processing. This might feel frustratingly slow, but you will get faster with practice. The immediate payoff will be the ease with which you can re-do data preparation when new data arrives. You can also re-use data preparation steps in the future for related projects. For very large data sets, data preparation may also include writing and saving scripts to obtain the data or subsets of the data from remote storage.
Some data cleaning tools, such as OpenRefine, provide a graphical user interface, but also automatically keep track of each step in the process. When tools like these or scripting is not feasible, it’s important to clearly document every manual action (what menu was used, what column was copied and pasted, what link was clicked, etc.). Often you can at least capture what action was taken, if not the complete why. For example, choosing a region of interest in an image is inherently interactive, but you can save the region chosen as a set of boundary coordinates.
How, when and why do you document?
As much as possible, always and to help you future self.
Use multiple tables as necessary, and use a unique identifier for every record
Raw data, even if tidy, is not necessarily complete. For example, the primary data table might hold the heart rate for individual subjects at rest and after a physical challenge, identified via a subject ID. Demographic variables, such as subject age and sex, are stored in a second table and will need to be brought in via merging or lookup. This will go more smoothly if subject ID is represented in a common format in both tables, e.g., always as “14025” versus “14,025” in one table and “014025” in another. It is generally wise to give each record or unit a unique, persistent key and to use the same names and codes when variables in two datasets refer to the same thing.
Submit data to a reputable DOI-issuing repository so that others can access and cite it
- Personal/lab web-site: not the best place to store your data long-term. These websites are not hosted long term. You can have a link to the repo, though.
- GitHub: in itself it is not proper for sharing your data as it can be modified. A snapshot of a Github repository can be stored in a service like Zenodo or the DAP and be issued a DOI.
- General data repo (i.e.: Zenodo, Data Dryad, etc.): good option to deposit data that does not fit in a specific repository. Best if the service is non-commerical, has long-termdata archival and issues DOIs, such as Zenodo. But DAP is preferred for CSIRO employees.
- Community specific repo (i.e.: ArrayExpress, NCBI, SRA, EGA, PRIDE, etc.): good option, if your research community has come up with a sustainable long-term repository for a certain data type.
- DAP: generally best option for any data sharing for CSIRO employees.
Your data is as much a product of your research as the papers you write, and just as likely to be useful to others (if not more so). Services such as the DAP can allow others to find your work, use it, and cite it; we discuss licensing in the episode on collaboration [04-collaboration]. Follow your research community’s standards for how to provide metadata. Note that there are two types of metadata: metadata about the dataset as a whole and metadata about the content within the dataset. If the audience is humans, write the metadata (the README file) for humans. If the audience includes automatic metadata harvesters, fill out the formal metadata and write a good README file for the humans [wickes2015].
What is a DOI?
- A digital object identifier is a persistent identifier or handle used to identify objects uniquely.
- Data with a persistent DOI can be found even when your lab website dies.
- doi-issuing repositories include: zenodo, figshare, dryad and the DAP.
- e.g. https://doi.org/10.25919/t1ad-8k76
Data management plans
Many universities and funders require researchers to complete a data management plan (DMP). A DMP is a document which outlines information about your research data and how it will be processed. Many funders provide basic templates for writing a DMP, along with guidelines on what information should be included but the main compoments of a DMP are:
- Information about your data
- Information about your metadata and data formats
- Information on how data can be accessed, shared and re-used
- Information on how data will be stored and managed, including long-term storage and maintenance after your project is complete
Discussion (2 minutes)
Aside from being a requirement, there are many benefits of writing a DMP to researchers. What sort of benefits do you think there are?
- Find and understand data easily
- Allows continuity of work when colleagues leave or join the lab
- It helps you consider issues about your data before they arise and come up with solutions
Writing your first data management plan can be a daunting task but your future self will thank you in the end. It’s best to speak to other members of your lab about any existing lab group or grant data management plans. If your lab group doesn’t have a data management plan, it may be helpful to work on it together to identify any major considerations. Often research institutions provide support for DMPs, e.g. through library services or a data steward.
More resources on data management plans are available at DMP online.
The CSIRO’s Research Data Planner can help you prepare a data management plan. It will be covered in detail next week.
What’s your next step in data management?
- Which recommendations above are most helpful for your current project? What could you try this week?
- Does your next project have a data management plan? Could you draft one?
Summary
Taken in order, the recommendations above will make it easier to keep track of your data and to work with it. Saving the raw data along with clear metadata, backed up, is your insurance policy. Creating analysis-friendly data, and recording all the steps used to process data, means that you and others can reproduce your analysis. Sharing your data via DOI-issuing repository allows others can access and cite it, which they will find easier if your data are analysis-friendly, clearly named, and well-documented.
These recommendations include explicitly creating and retaining of intermediate data files at intermediate steps of the analysis, with increasing levels of cleanliness and task-specificity. Saving intermediate files makes it easy to re-run parts of a data analysis pipeline, which in turn makes it less onerous to revisit and improve specific data processing tasks. Breaking a lengthy analysis workflow into modular parts makes it easier to understand, share, describe, and modify.
Modifying and sharing your data analysis is only possible if you still have the raw data: back up your data!!!
Attribution
This episode was adapted from and includes material from Wilson et al. Good Enough Practices for Scientific Computing.
Key Points
- Raw data is the data as originally generated – it should be kept read-only
- Raw data has to be backed up in more than one location
- Create the data you wished you have received
- Keeping track of your actions is a key part of data management
- The Digital object identifiers (DOIs) is a unique identifier that permanently identifies data and makes it findable
- Finding a repository tailored to your data is key to making it findable and accessible by the broader community
Content from Code and Software
Last updated on 2024-01-28 | Edit this page
Estimated time: 40 minutes
Overview
Questions
- What is research code and software?
- What can you do to make code usable and reusable?
- What are the characteristics of readable code?
Objectives
- Describe what is research software and its purposes
- Decompose a workflow into identifiable components
- Know when to separate a script into several functions
- Write code that is easy to run by others by including all dependencies, requirements, documentation, and examples
What your future self may think…

The twitter thread illustrates a real example of research software problems, from Prof. Neil Ferguson. In that case, the research software written for one purpose was suddenly in high demand, for important public health reasons, over a decade later. And that software was hard for others to use.
Smaller-scale versions of this problem are more common:
- you want to re-run a data analysis that you did six months ago
- a new post-doc starts working on a related project and needs to adapt your analysis to a new dataset
- you publish a paper, and a masters student from the other side of the world emails you to reproduce the results for their project
What is research code and software?
There are many different shapes and sizes of research software:
- Any code that runs in order to process your research data.
- A record of all the steps used to process your data (scripts and workflow such data analysis are software).
- R, Python, MATLAB, unix shell, OpenRefine, ImageJ, etc. are all scriptable. So are Microsoft Excel macros.
- Standalone programs or scripts that do particular research tasks are also research software.
There are extended discussions about research software at the Software Sustainability Institute.
Potential problems writing code
Discussion
What can go wrong when trying to reuse research code?
- I don’t remember what this code does
- I don’t remember why I made this choice
- This code doesn’t work any more
- This code doesn’t work on an updated version of my dataset
- This code doesn’t work on a different computing system
- I’m not sure if this calculation is correct
If you or your group are creating ten thousands lines of code for use by hundreds of people you have never met, you are doing software engineering. If you’re writing a few dozen lines now and again, and are probably going to be its only user, you may not be doing engineering, but you can still make things easier on yourself by adopting a few key engineering practices. What’s more, adopting these practices will make it easier for other people and your future self to understand and (re)use your code.
The core realization in these practices is that readable, reusable, and testable are all side effects of writing modular code, i.e., of building programs out of short, single-purpose functions with clearly-defined inputs and outputs [hunt1999]. Much has been written on this topic, and this section focuses on practices that best balance ease of use with benefit for you and collaborators.
Programs themselves are modular, and can be written in scripts that run a clearly-defined set of functions on defined inputs.
Place a brief explanatory comment at the start of every script or program
Short is fine; always include at least one example of how the program is used. Remember, a good example is worth a thousand words. Where possible, the comment should also indicate reasonable values for parameters like in this example.
Synthesize image files for testing circularity estimation algorithm.
Usage: make_images.py -f fuzzing -n flaws -o output -s seed -v -w size
where:
-f fuzzing = fuzzing range of blobs (typically 0.0-0.2)
-n flaws = p(success) for geometric distribution of # flaws/sample (e.g. 0.5-0.8)
-o output = name of output file
-s seed = random number generator seed (large integer)
-v = verbose
-w size = image width/height in pixels (typically 480-800)
-h = show help messageThe reader doesn’t need to know what all these words mean for a comment to be useful. This comment tells the reader what words they need to look up: fuzzing, blobs, and so on.
Writing helpful explanatory comments
Multiple Choice
An example function GetData reads in data files of a
particular type. Which of the following should be included in an
explanatory comment for this function?
- file type - it is good to know what the function is tailored to process
- output type - it is good to know how to integrate a function in a workflow
- data columns or other properties - it is good to know the internal structure of the data
- expected file path / address (for example a specific directory or web address) - it is good to know the origin to evaluate the validity of the data
Decompose programs into functions
A function is a reusable section of software that can be treated as a black box by the rest of the program. This is like the way we combine actions in everyday life. Suppose that it is teatime. You could get a teabag, put the teabag in a mug, boil the kettle, pour the boiling water into the mug, wait 3 minutes for the tea to brew, remove the teabag, and add milk if desired. It is much easier to think of this as a single function, “make a cup of tea”.
Software programming languages also allow you to combine many steps into a single function. The syntax for creating functions depends on programming language, but generally you:
- name the function
- list its input parameters
- describe what information it produces
- write some lines of code that produce the desired output.
Good functions should have only one main task: for example, “make a cup of tea” does not also specify how to make a sandwich. Functions can also be built up from other functions: for example, “boil the kettle” involves checking if there is water in the kettle, filling the kettle if not, and then turning the kettle on. Having one main task means that functions should take no more than five or six input parameters and should not reference outside information. Functions should be no more than one page (about 60 lines) long: you should be able to see the entire function in a standard (~10pt) font on a laptop screen. If your function grows larger than this, it is usually best to break that up into simpler functions.
The key motivation here is to fit the program into the most limited memory of all: ours. Human short-term memory is famously incapable of holding more than about seven items at once [miller1956]. If we are to understand what our software is doing, we must break it into chunks that obey this limit, then create programs by combining these chunks. Putting code into functions also makes it easier to test and troubleshoot when things go wrong.
Pseudocode is a plain language description of code or analysis steps. Writing pseudocode can be useful to think through the logic of your analysis, and how to decompose it into functions.
The “make a cup of tea” example above might look like this:
make_cup_of_tea = function(sugar, milk)
if kettle is not full
fill kettle
boil kettle
put teabag in cup
add water from kettle to cup
wait 2 minutes
if sugar is true
add sugar to cup
if milk is true
add milk to cup
stir with spoon
return cupUsing pseudocode
In this scenario, you’re managing fruit production on a set of islands. You have written a pseudocode function that tells you how to count how much fruit of a particular type is available to harvest on a given island.
count_fruit_on_island = function(fruit type, island)
total fruit = 0
for every tree of fruit type on the island
total fruit = total fruit + number of fruit on tree
end for loop
return total fruitWrite a pseudocode command to call this function to count how many coconuts there are on Sam’s island.
Write a pseudocode for loop like the one above that uses this function to count all the cherries on every island.
To count Sam’s island’s coconuts:
sams coconuts = count_fruit_on_island(coconuts, Sam's island)To count all the cherries on every island:
total cherries = 0
for every island
total cherries = total cherries + count_fruit_on_island(cherries, island)
end for loop
print "There are " + total cherries + " cherries on all the islands"Be ruthless about eliminating duplication
Write and re-use functions instead of copying and pasting code, and
use data structures like lists instead of creating many closely-related
variables, e.g. create score = (1, 2, 3) rather than
score1, score2, and score3.
Also look for well-maintained libraries that already do what you’re trying to do. All programming languages have libraries that you can import and use in your code. This is code that people have already written and made available for distribution that have a particular function. For instance, there are libraries for statistics, modeling, mapping and many more. Many languages catalog the libraries in a centralized source, for instance R has CRAN, Python has PyPI, and so on. So always search for well-maintained software libraries that do what you need before writing new code yourself, but test libraries before relying on them.
Give functions and variables meaningful names
Meaningful names for functions and variables document their purpose
and make the program generally easy to read. As a rule of thumb, the
greater the scope of a variable, the more informative its name should
be: while it’s acceptable to call the counter variable in a loop
i or j, things that are re-used often, such as
the major data structures in a program should not have
one-letter names.
Name that function
An example function is defined in the format
functionName (variableName) This function cubes every third
number in a sequence. What are the most meaningful names for
functionName and variableName? Choose one from
each of the following sections:
functionName
- processFunction
- computeCubesOfThird
- cubeEveryThirdNumberInASequence
- cubeEachThird
- 3rdCubed
variableName
- arrayOfNumbersToBeCubed
- input
- numericSequence
- S
functionName
- processFunction - incorrect, too vague
- computeCubesOfThird - incorrect, doesn’t imply every third in sequence
- cubeEveryThirdNumberInASequence - maybe, but too long
- cubeEachThird - correct, short and includes information on the data and calculation performed
- 3rdCubed - incorrect, bad practice to put a number at the beginning of a function name (not allowed by some programming languages)
variableName
- arrayOfNumbersToBeCubed - maybe, but too long
- input - incorrect, too vague
- numericSequence - correct, short and included information about the type of input
- S - incorrect, too vague
Language style guides
Remember to follow each language’s conventions for names, such as
net_charge for Python and NetCharge for Java.
These conventions are often described in “style guides”, and can even be
checked automatically.
Callout
Tab Completion
Almost all modern text editors provide tab completion, so that typing the first part of a variable name and then pressing the tab key inserts the completed name of the variable. Employing this means that meaningful longer variable names are no harder to type than terse abbreviations.
Make dependencies and requirements explicit.
This is usually done on a per-project rather than per-program basis,
i.e., by adding a file called something like
requirements.txt to the root directory of the project, or
by adding a “Getting Started” section to the README
file.
Do not comment and uncomment sections of code to control a program’s behavior
This is error prone and makes it difficult or impossible to automate
analyses. Instead, put if/else statements in the program to control what
it does, and use input arguments on the command line to select
particular behaviour. For example, including the input argument
--option and corresponding if/else statements to control
running an optional piece of the program. Remember to use descriptive
names for input arguments.
Provide a simple example or test data set
Users (including yourself) can run your program on this set to determine whether it is working and whether it gives a known correct output for a simple known input. Such a test is particularly helpful when supposedly-innocent changes are being made to the program, or when it has to run on several different machines, e.g., the developer’s laptop and the department’s cluster. This type of test is called an integration test.
Code can be managed like data
Your code is like your data and also needs to be managed, backed up, and shared.
Your software is as much a product of your research as your papers, and should be as easy for people to credit. Submit code to a reputable DOI-issuing repository, just as you do with data. DOIs for software are provided by Figshare and Zenodo, for example. Both Figshare and Zenodo integrate directly with GitHub.
Attribution
This episode was adapted from and includes material from Wilson et al. Good Enough Practices for Scientific Computing.
Key Points
- Any code that runs on your research data is research software
- Write your code to be read by other people, including future you
- Decompose your code into modules: scripts and functions, with meaningful names
- Be explicit about requirements and dependencies such as input files, arguments and expected behaviour
Content from Collaboration
Last updated on 2024-01-29 | Edit this page
Estimated time: 37 minutes
Overview
Questions
- What do collaborators need to know to contribute to my project?
- How can documentation make my project more efficient?
- What is a license and does my project need one?
Objectives
- Facilitate contributions from present and future collaborators
- Learn to treat every project as a collaborative project
- Describe a project in a README file
- Understand what software licenses are and how they might apply to your project
You may start working on projects by yourself or with a small group of collaborators you already know, but you should design it to make it easy for new collaborators to join. These collaborators might be new grad students or postdocs in the lab, or they might be you returning to a project that has been idle for some time. As summarized in [steinmacher2015], you want to make it easy for people to set up a local workspace so that they can contribute, help them find tasks so that they know what to contribute, and make the contribution process clear so that they know how to contribute. You also want to make it easy for people to give you credit for your work.
Collaboration opportunities and challenges
Discussion
- How does collaboration help in scientific computing?
- What goes wrong with collaboration?
- How can you prepare to collaborate?
How collaboration can help:
- Collaboration brings other ideas and perspectives on your project
- Describing your project to (potential) collaborators can help to focus the project
- Thinking about other people helps you to return to your project later
What can go wrong with collaboration:
- People can be confused about:
- Goals: what are we trying to do?
- Process: what tools will we use, how will we do it?
- Responsibilities: whose job is it to do this thing?
- Credit: how are contributions going to be recognized?
- Data: how do we share sensitive data?
- Timelines: when will people finish their tasks?
How to prepare for collaboration:
- Document important things
- Decide on goals and a way of working (process)
- Clarify the scope and audience of your project
- Highlight outstanding issues
Create an overview of your project
Written documentation is essential for collaboration. Future you will forget things, and your collaborators will not know them in the first place. An overview document can collect the most important information about your project, and act as a signpost. The overview is usually the first thing people read about your project, so it is often called a “README”. The README has two jobs: describing the contents of the project, and explaining how to interact with the project.
Create a short file in the project’s home directory that explains the
purpose of the project. This file (generally called README,
README.txt, or something similar) should contain :
- The project’s title
- A brief description
- Up-to-date contact information
- An example or two of how to run the most important tasks
- Overview of folder structure
Describe how to contribute to the project
Because the README is usually the first thing users and collaborators on your project will look at, make it explicit how you want people to engage with the project. If you are looking for more contributors, make it explicit that you welcome contributors and point them to the license (more below) and ways they can help.
A separate CONTRIBUTING file can also describe what
people need to do in order to get the project going and use or
contribute to it:
- Dependencies that need to be installed
- Tests that can be run to ensure that software has been installed correctly
- Guidelines or checklists that your project adheres to.
This information is very helpful and will be forgotten over time unless it’s documented inside the project.
Comparing README files
Here is a README file for a data project and one for a software
project. What useful and important information is present, and what is
missing? Data
Project README
Software Project
README
This Data Project README:
- Contains a DOI
- Describes the purpose of the code and links to a related paper
- Describes the project structure
- Includes a license
- DOES NOT contain requirements
- DOES NOT include a working example
- DOES NOT include a explicit list of authors (can be inferred from paper though)
This Software Project README:
- Describes the purpose of the code
- Describes the requirements
- Includes instructions for various type of users
- Describes how to contribute
- Includes a working example
- Includes a license
- DOES NOT include an explicit DOI
- DOES NOT describe the project structure
Decide on communication strategies
Make explicit decisions about (and publicize where appropriate) how members of the project will communicate with each other and with external users / collaborators. This includes the location and technology for email lists, chat channels, voice / video conferencing, documentation, and meeting notes, as well as which of these channels will be public or private.
Supported tools within CSIRO include Confluence, for wiki-style shared documentation, and Microsoft Teams, for online discussions, video conferencing, file sharing and more.
Not supported, but worth knowing of is Miro, which allows creating a shared, online whiteboard, where multiple people may be building up notes, diagrams, graphs, etc., at the same time, with quite powerful tools.
Collaborations with sensitive data
If you determine that your project will include work with sensitive data, it is important to agree with collaborators on how and where the data will be stored, as well as what the mechanisms for sharing the data will be and who is ultimately responsible for ensuring these are followed.
Create a shared “to-do” list
Organising a structured to-do list of tasks still to complete and overall project work plan can be really useful whether collaborating with others or just with your future self. There are many options and tools available for how to do this.
If your project is centered around a Git-tracked repository, it could be as simple as updating a text file like
notes.txtortodo.txt.Or make use of the ability to track “issues” on BitBucket or GitHub. The “issues” feature on online Git repositories allows you or others to describe work that needs to be done (often used on public repositories for reporting bugs in software), create and follow discussions about the issues/tasks, and link the closing of the issues to commits/pull-requests. E.g.: Issues for original version of this lesson
Microsoft Teams ‘Tasks’. Teams now lets you add a ‘Tasks’ app/tab to a team space, which then lets create a to-do list, assign tasks to people, and lets you track and view the tasks in various ways.
Jira is another tool supported and deployed in CSIRO. Developed by Australian software company Atlassian, it allows tracking of to-do tasks/issues and sub-tasks, lets you assign tasks to people, and lets you track and view tasks in the context of worflows, timelines, and “board” visualisations, such as the “Kanban board”. Jira can directly integrate with both BitBucket and Confluence, with Jira tasks able to be linked, referenced and tracked in each. Jira can be a very powerful tool if fully embraced, but can also be a bit clunky to starting out.
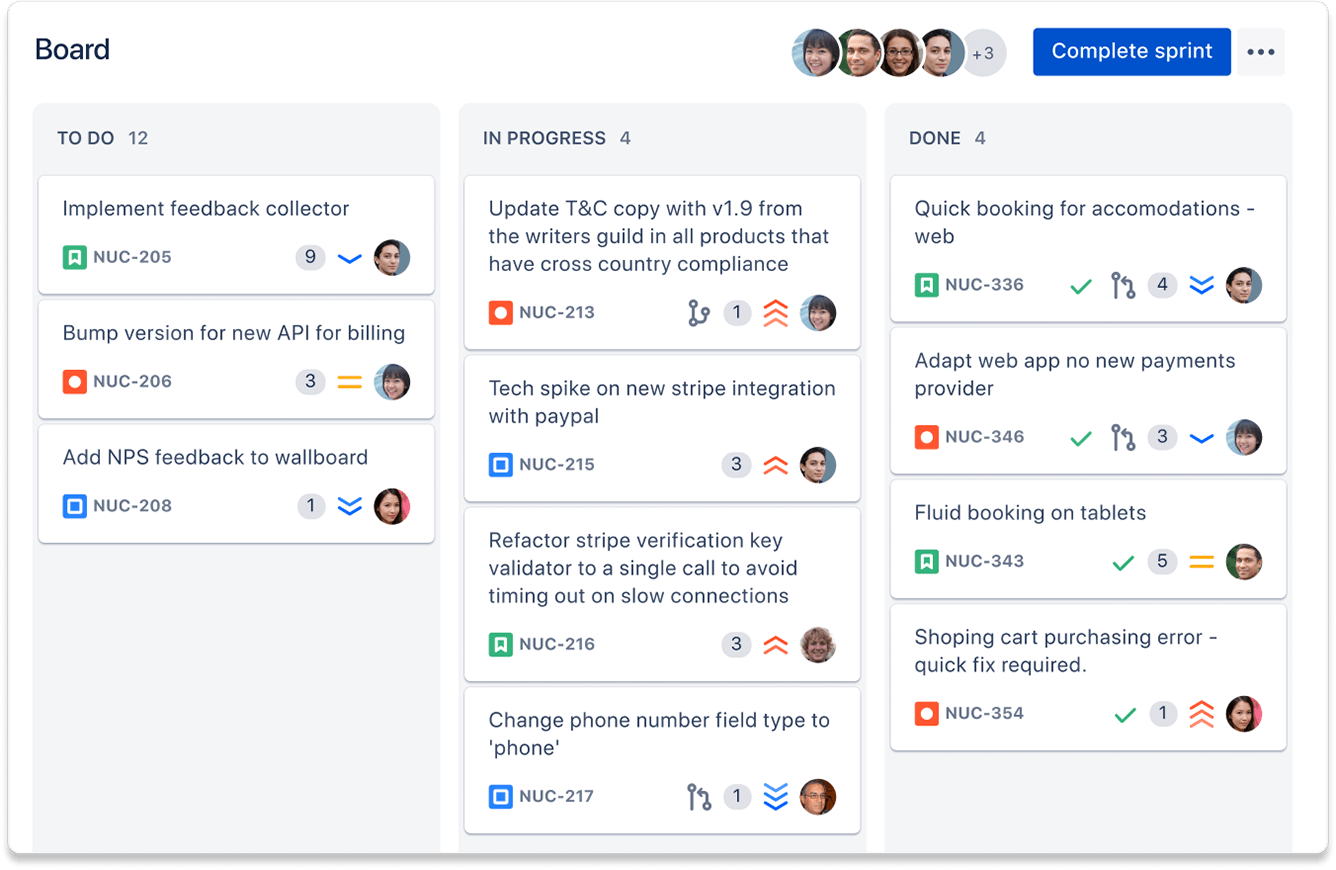 A Kanban board example, from
Jira’s
website.
A Kanban board example, from
Jira’s
website.
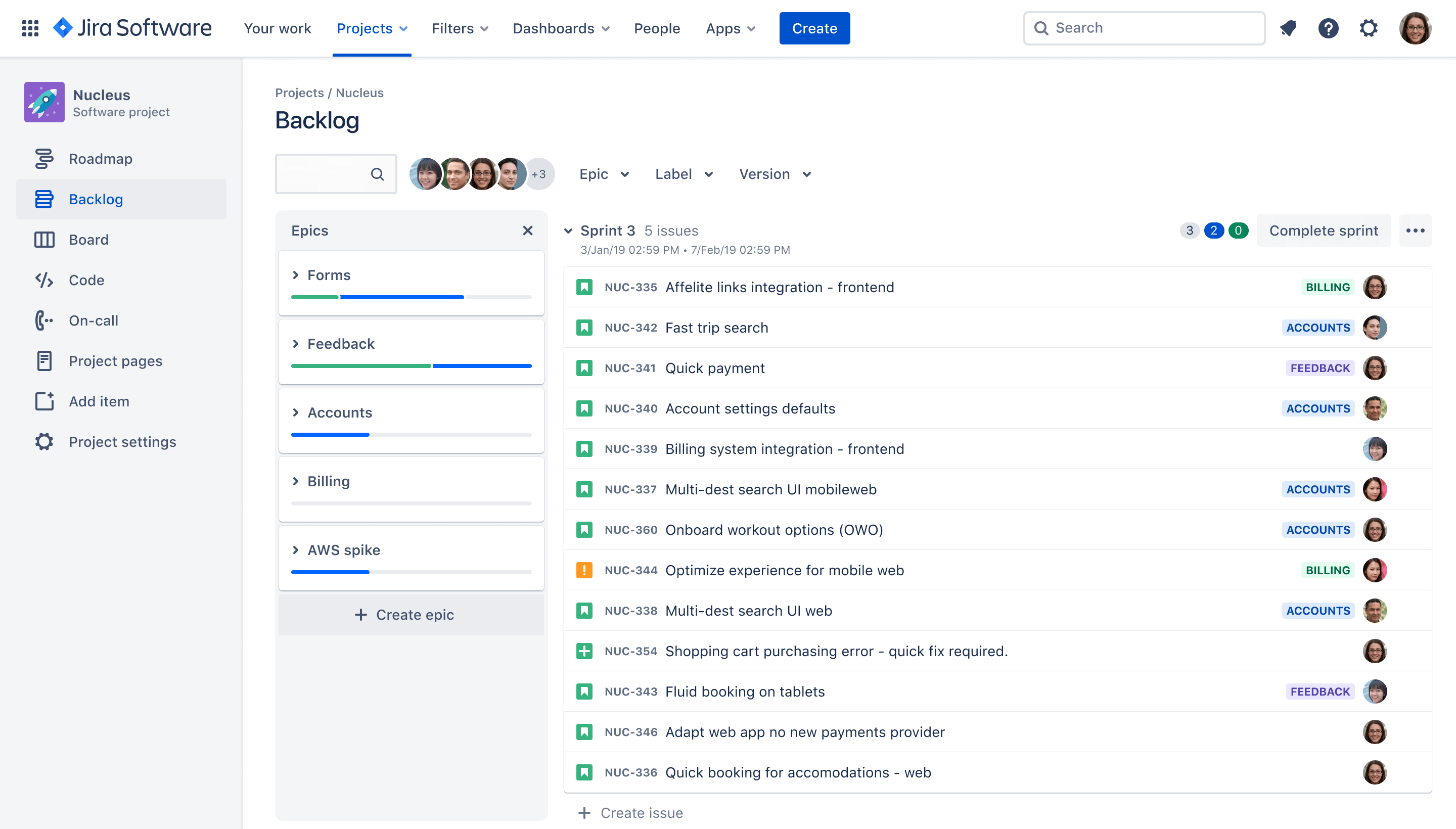 A “backlog” view example, from Jira’s
website.
A “backlog” view example, from Jira’s
website.
Make the license explicit
What is a licence?
- Specifies allowable copying and reuse
- Without a licence, people cannot legally reuse your code or data
- Different options for different goals and funder requirements (Apache, MIT, CC, …)
- For example, this lesson is reusable with attribution under a Creative Commons Attribution (CC BY) 4.0 licence
- Applies to all material in a project, e.g. data, text and code
Have a LICENSE file in the project’s home directory that
clearly states what license(s) apply to the project’s software, data,
and manuscripts. Lack of an explicit license does not mean there isn’t
one; rather, it implies the author is keeping all rights and others are
not allowed to re-use or modify the material. A project that consists of
data and text may benefit from a different license to a project
consisting primarily of code.
IM&T can help with advising on suitable licenses.
The original authors of this lesson recommend Creative Commons licenses for data and text, either CC-0 (the “No Rights Reserved” license) or CC-BY (the “Attribution” license, which permits sharing and reuse but requires people to give appropriate credit to the creators). For software, we recommend a permissive open source license such as the MIT, BSD, or Apache license [laurent2004]. A useful resource to compare different licenses is available at tldrlegal. More advice for how to use licences for research data is available at openaire.
What Not To Do
We (the original authors) recommend against the “no commercial use” variations of the Creative Commons licenses because they may impede some forms of re-use. For example, if a researcher in a developing country is being paid by her government to compile a public health report, she will be unable to include your data if the license says “non-commercial”. We recommend permissive software licenses rather than the GNU General Public License (GPL) because it is easier to integrate permissively-licensed software into other projects, see chapter three in [laurent2004].
Make the project citable
A CITATION file describes how to cite your project as a
whole, and where to find (and how to cite) any data sets, code, figures,
and other artifacts that have their own DOIs. The example below shows
the CITATION file for the Ecodata Retriever; for
an example of a more detailed CITATION file, see the one
for the khmer
project.
Please cite this work as:
Morris, B.D. and E.P. White. 2013. "The EcoData Retriever:
improving access to existing ecological data." PLOS ONE 8:e65848.
http://doi.org/doi:10.1371/journal.pone.0065848More recently a standard for citation files has been developed in the form of CFF; Citation File Format. Often saved in a CITATION.cff file, this format was proposed specifically for the purpose of holding expected information as a standardised set that was both human and machine readable. E.g.:
cff-version: 1.2.0
message: "If you use this software, please cite it as below."
authors:
- family-names: Druskat
given-names: Stephan
orcid: https://orcid.org/1234-5678-9101-1121
title: "My Research Software"
version: 2.0.4
doi: 10.5281/zenodo.1234
date-released: 2021-08-11More information on CFF is available here: citation-file-format.github.io
Recommended resources
Attribution
This episode was adapted from and includes material from Wilson et al. Good Enough Practices for Scientific Computing.
Key Points
- Create an overview of your project
- Create a shared “to-do” list
- Decide on communication strategies
- Make the license explicit
- Make the project citable
Content from Project Organization
Last updated on 2024-01-29 | Edit this page
Estimated time: 25 minutes
Overview
Questions
- How should I name my files?
- How does folder organization help me
Objectives
- Understand elements of good naming strategy
- Evaluate pros and cons of different project organizations
Organizing the files that make up a project in a modular, logical, and consistent directory structure will help you and others keep track of them.
Project organisation problems
Discuss what can go wrong with project organisation:
- Struggling to find the code that creates a particular figure
- Hating to look at or even think about your project because of how badly organised it is
README files are magic
You look at a directory (or project), you read it, and it tells you what you need to know
… as long as you keep it updated!
Put each project in its own directory, which is named after the project
Similar to deciding when a chunk of code should be made a function, the ultimate goal of dividing research into distinct projects is to help you and others best understand your work. Some researchers create a separate project for each manuscript they are working on, while others group all research on a common theme, data set, or algorithm into a single project.
As a rule of thumb, divide work into projects or modules based on the overlap in data and code files. If two research efforts share no data or code, they will probably be easiest to manage independently. If they share more than half of their data and code, they are probably best managed together, while if you are building tools that are used in several projects, the common code should probably be in a project of its own.
Projects do often require their own organizational model. The below recommendations on how you can structure data, code, analysis outputs and other files, are drawn primarily from [noble2009, gentzkow2014]. Other structures to consider are:
The important concepts are that is useful to organize the project in modules by the types of files and that consistent planning and good names help you effectively find and use things later. Your lab or organization may have a template to use.
Put text documents associated with the project in the
doc directory.
This includes files for manuscripts, documentation for source code, and/or an electronic lab notebook recording your experiments. Subdirectories may be created for these different classes of files in large projects.
Put raw data and metadata in a data directory, and
files generated during cleanup and analysis in a results
directory
When we refer to “generated files”, this includes intermediate results, such as cleaned data sets or simulated data, as well as final results such as figures and tables.
The results directory will usually require
additional subdirectories for all but the simplest projects.
Intermediate files such as cleaned data, statistical tables, and final
publication-ready figures or tables should be separated clearly by file
naming conventions or placed into different subdirectories; those
belonging to different papers or other publications should be grouped
together. Similarly, the data directory might require
subdirectories to organize raw data based on time, method of collection,
or other metadata most relevant to your analysis.
Put project source code in the src directory
src contains all of the code written for the project.
This includes programs written in interpreted languages such as R or
Python; those written compiled languages like Fortran, C++, or Java; as
well as shell scripts, snippets of SQL used to pull information from
databases; and other code needed to regenerate the results.
This directory may contain two conceptually distinct types of files that should be distinguished either by clear file names or by additional subdirectories. The first type are files or groups of files that perform the core analysis of the research, such as data cleaning or statistical analyses. These files can be thought of as the “scientific guts” of the project.
Another type of file that might go in src is
controller/driver/workflow scripts that contain all the analysis steps
of a project from start to finish, with particular parameters and data
input/output commands. A controller script for a simple project, for
example, may read a raw data table, import and apply several cleanup and
analysis functions from the other files in this directory, and create
and save a numeric result. For a small project with one main output, a
single controller script could be placed in the main src
directory and distinguished clearly by a name such as “runall”. The
short example below is typical of scripts of this kind; note how it uses
one variable, TEMP_DIR, to avoid repeating the name of a
particular directory four times.
TEMP_DIR = ./temp_zip_files
echo "Packaging zip files required by analysis tool..."
mkdir $(TEMP_DIR)
./src/make-zip-files.py $(TEMP_DIR) *.dat
echo "Analyzing..."
./bin/sqr_mean_analyze -i $(TEMP_DIR) -b "temp"
echo "Cleaning up..."
rm -rf $(TEMP_DIR)Callout
Important note: Don’t place information specific to
your own computer/system or self in these types of files, especially if
they are being Git-tracked. Use relative paths instead of full paths
where possible (e.g. input as ../data/ rather than
/home/xyz123/project/data). Don’t include any passwords or
keys.
If personal or system-specific information is required for your
workflow, then make use of locally set environment variables and/or
git-ignored files and then document how to set up these inputs again for
anyone (or future self) re-using your work.
Put compiled programs in the bin directory
A directory named bin is usually used to contain
executable programs compiled from code in the src
directory. Projects that do not have any will not require
bin.
Scripts vs. Programs
We use the term “script” to mean “something that is executed directly as-is”, and “program” to mean “something that is explicitly compiled before being used”. The distinction is more one of degree than kind—libraries written in Python are actually compiled to bytecode as they are loaded, for example—so one other way to think of it is “things that are edited directly” and “things that are not”.
External Scripts
If
srcis for human-readable source code, andbinis for compiled binaries, where should projects put scripts that are executed directly—particularly ones that are brought in from outside the project? On the one hand, these are written in the same languages as the project-specific scripts insrc; on the other, they are executable, like the programs inbin. The answer is that it doesn’t matter, as long as each team’s projects follow the same rule. As with many of our other recommendations, consistency and predictability are more important than hair-splitting.
Name all files to reflect their content or function.
For example, use names such as bird_count_table.csv,
manuscript.md, or sightings_analysis.py. Do
not use sequential numbers (e.g., result1.csv,
result2.csv) or a location in a final manuscript (e.g.,
fig_3_a.png), since those numbers will almost certainly
change as the project evolves.
File names should be:
- Machine readable
- Human readable
- Descriptive of their contents
- Optional: Consistent
- Optional: Play well with default ordering
Example
The diagram below provides a concrete example of how a simple project might be organized following these recommendations:
.
|-- CITATION.cff
|-- README.md
|-- LICENSE.md
|-- requirements.txt
|-- data
| -- birds_count_table.csv
|-- doc
| -- notebook.md
| -- manuscript.md
| -- changelog.txt
|-- results
| -- summarized_results.csv
|-- src
| -- sightings_analysis.py
| -- runall.pyThe root directory contains a README file that provides
an overview of the project as a whole, a CITATION file that
explains how to reference it, and a LICENSE file that
states the licensing. The requirements.txt file lists the
software that is required to run the data analysis. The
data directory contains a single CSV file with tabular data
on bird counts (machine-readable metadata could also be included here).
The src directory contains
sightings_analysis.py, a Python file containing functions
to summarize the tabular data, and a controller script
runall.py that loads the data table, applies functions
imported from sightings_analysis.py, and saves a table of
summarized results in the results directory.
This project doesn’t have a bin directory, since it does
not rely on any compiled software. The doc directory
contains two text files written in Markdown, one containing a running
lab notebook describing various ideas for the project and how these were
implemented and the other containing a running draft of a manuscript
describing the project findings.
Naming and sorting (5 minutes)
Have a look at the example files from a project, similar to the one from the previous metadata episode.
All the files have been sorted by name and demonstrate consequences of different naming strategies.
For your information, to encode experimental details the following conventions were taken:
- phyB/phyA are sample genotypes (that is, which gene is mutated)
- sXX is the sample number
- LD/SD are different light conditions (long or short day)
- on/off are different media (on sucrose, off sucrose)
- measurement date
- other details are timepoint and raw or normalized data
2020-07-14_s12_phyB_on_SD_t04.raw.csv
2020-07-14_s1_phyA_on_LD_t05.raw.csv
2020-07-14_s2_phyB_on_SD_t11.raw.csv
2020-08-12_s03_phyA_on_LD_t03.raw.csv
2020-08-12_s12_phyB_on_LD_t01.raw.csv
2020-08-13_s01_phyB_on_SD_t02.raw.csv
2020-7-12_s2_phyB_on_SD_t01.raw.csv
AUG-13_phyB_on_LD_s1_t11.raw.csv
JUL-31_phyB_on_LD_s1_t03.raw.csv
LD_phyA_off_t04_2020-08-12.norm.csv
LD_phyA_on_t04_2020-07-14.norm.csv
LD_phyB_off_t04_2020-08-12.norm.csv
LD_phyB_on_t04_2020-07-14.norm.csv
SD_phyB_off_t04_2020-08-13.norm.csv
SD_phyB_on_t04_2020-07-12.norm.csv
SD_phya_off_t04_2020-08-13.norm.csv
SD_phya_ons_t04_2020-07-12.norm.csv
ld_phyA_ons_t04_2020-08-12.norm.csv - What are the problems with having the date first?
- How do different date formats behave once sorted?
- Can you tell the importance of a leading 0 (zeros)?
- Is it equally easy to find all data from LD conditions as ON media?
- Can you spot the problem when using different cases (upper/lower)?
- Do you see benefits of keeping consistent lengths of the naming conventions?
- Do you see what happens when you mix conventions?
- Using dates up front makes it difficult to quickly find data for particular conditions or genotypes. It also masks the “logical” order of samples or timepoints.
- Named months break the “expected” sorting, same as dates without leading 0
- Without leading zeros, ‘s12’ appear before s1 and s2
- the first (and second) part of the name are easiest to spot
- the last file is also from LD conditions, but appears after SD, same with ‘phya’ genotypes
- the last 3 file names are easiest to read as all parts appear on top of each other due to the same 3 letter-length codes ons and off
- The lack of consistency makes it very difficult to get data from related samples/conditions.
Some helpful organisation tools
- Integrated Development Environments (IDEs) combine many features to
write code and organise projects:
- PyCharm for python
- RStudio for R
- VSCode for python and other languages, etc.
- Notebooks collect data, code, results, thinking in single documents:
- jupyter
- R markdown
- quarto, etc.
- Cookie Cutter project templates for reproducible science
Attribution
This episode was adapted from and includes material from Wilson et al. Good Enough Practices for Scientific Computing.
Some content was adapted from FAIR in Biological Practice episode on files and organisation. That material gives a slightly different and also useful perspective.
Key Points
- A good file name suggests the file content
- Good project organization saves you time
Content from Keeping Track of Changes
Last updated on 2024-01-29 | Edit this page
Estimated time: 40 minutes
Overview
Questions
- How do I make changes to a project without losing or breaking things?
- Why does GitHub exist?
Objectives
- List common problems with introducing changes to files without tracking
- Understand good practices in tracking changes
- Write a good change description
Problems with change
Which of this issues can you relate to?
- I have fifteen versions of this file and I don’t know which is which
- I can’t remake this figure from last year
- I modified my code and something apparently unrelated does not work anymore
- I have several copies of the same directory because I’m worried about breaking something
- Somebody duplicated a record in a shared file with samples
- You remember seeing a data file but cannot find it anymore: is it deleted ? Moved away ?
- I tried multiple analysis and I don’t remember which one I chose to generate my output data
- I have to merge changes to a paper from mails with collaborators
- I accidently deleted a part of my work
- I came to an old project and forgot where I left it
- I have trouble to find the source of a mistake in an experiment
- My directory is polluted with a lot of unused/temporary/old folders because I’m afraid of losing something important
- I made a lot of changes to my paper but only want to bring back one of paragraph
Keeping track of changes that you or your collaborators make to data and software is a critical part of research. Being able to reference or retrieve a specific version of the entire project aids in reproducibility for you leading up to publication, when responding to reviewer comments, and when providing supporting information for reviewers, editors, and readers.
We believe that the best tools for tracking changes are the version control systems that are used in software development, such as Git, Mercurial, and Subversion. They keep track of what was changed in a file when and by whom, and synchronize changes to a central server so that many users can manage changes to the same set of files.
While these version control tools make tracking changes easier, they can have a steep learning curve. So, we provide two sets of recommendations:
- a systematic manual approach for managing changes and
- version control in its full glory,
and you can use the first while working towards the second, or just jump in to version control.
Whatever system you chose, we recommend that you:
Back up (almost) everything created by a human being as soon as it is created
This includes scripts and programs of all kinds, software packages that your project depends on, and documentation. A few exceptions to this rule are discussed below.
Keep changes small
Each change should not be so large as to make the change tracking irrelevant. For example, a single change such as “Revise script file” that adds or changes several hundred lines is likely too large, as it will not allow changes to different components of an analysis to be investigated separately. Similarly, changes should not be broken up into pieces that are too small. As a rule of thumb, a good size for a single change is a group of edits that you could imagine wanting to undo in one step at some point in the future.
Share changes frequently
Everyone working on the project should share and incorporate changes from others on a regular basis. Do not allow individual investigator’s versions of the project repository to drift apart, as the effort required to merge differences goes up faster than the size of the difference. This is particularly important for the manual versioning procedure described below, which does not provide any assistance for merging simultaneous, possibly conflicting, changes.
Create, maintain, and use a checklist for saving and sharing changes to the project
The list should include writing log messages that clearly explain any changes, the size and content of individual changes, style guidelines for code, updating to-do lists, and bans on committing half-done work or broken code. See [gawande2011] for more on the proven value of checklists.
Store each project in a folder that is mirrored off the researcher’s working machine
This may include:
- using a shared system such as a (institutional) cloud or shared drive, or
- a remote version control repository such as GitHub.
Synchronize that folder at least daily. It may take a few minutes, but that time is repaid the moment a laptop is stolen or its hard drive fails.
How to document a change
A good entry that documents changes should contain:
- Date of the change
- Author of the change
- List of affected files
- A short description of the nature of the introduced changes AND/OR motivation behind the change.
Examples of the descriptions are:
Added flow cytometry data for the control and starvation stressed samples
Updated matplot library to version 3.4.3 and regenerated figures
Added pane with protein localization to the Figure 3 and its discussion in the text
Reverted to the previous version of the abstract text as the manuscript reached word limits
Cleaned the strain inventory: Recent freezer cleaning and ordering indicated a lot of problem with the strains data. The missing physical samples were removed from the table, the duplicated ids are marked for checking with PCR. The antibiotic resistance were moved from phenotype description to its own column.
New regulation heatmap: As suggested by Will I used the normalization and variance stabilization procedure from Hafemeister et al prior to clustering and heatmap generation
The larger the project (measured either in: collaborators, file numbers, or workflow complexity) the more detailed the change description should be. While your personal project can get away with one liner descriptions, the largest projects should always contain information about motivation behind the change and what are the consequences.
Manual Versioning
Our first suggested approach, in which everything is done by hand, has two additional parts:
-
Add a file called
CHANGELOG.txtto the project’sdocssubfolder, and make dated notes about changes to the project in this file in reverse chronological order (i.e., most recent first). This file is the equivalent of a lab notebook, and should contain entries like those shown below.
## 2016-04-08
* Switched to cubic interpolation as default.
* Moved question about family's TB history to end of questionnaire.
## 2016-04-06
* Added option for cubic interpolation.
* Removed question about staph exposure (can be inferred from blood test results).- Copy the entire project whenever a significant change has been made (i.e., one that materially affects the results), and store that copy in a sub-folder whose name reflects the date in the area that’s being synchronized. This approach results in projects being organized as shown below:
.
|-- project_name
| -- current
| -- ...project content as described earlier...
| -- 2016-03-01
| -- ...content of 'current' on Mar 1, 2016
| -- 2016-02-19
| -- ...content of 'current' on Feb 19, 2016Here, the project_name folder is mapped to external
storage (such as Dropbox), current is where development is
done, and other folders within project_name are old
versions.
Data is Cheap, Time is Expensive
Copying everything like this may seem wasteful, since many files won’t have changed, but consider: a terabyte hard drive costs about \$50, which means that 50 GByte costs less than \$5. Provided large data files are kept out of the backed-up area (discussed below), this approach costs less than the time it would take to select files by hand for copying.
This manual procedure satisfies the requirements outlined above without needing any new tools. If multiple researchers are working on the same project, though, they will need to coordinate so that only a single person is working on specific files at any time. In particular, they may wish to create one change log file per contributor, and to merge those files whenever a backup copy is made.
Version Control Systems
What the manual process described above requires most is self-discipline. The version control tools that underpin our second approach—the one we use in our own projects–don’t just accelerate the manual process: they also automate some steps while enforcing others, and thereby require less self-discipline for more reliable results.
It’s hard to know what version control tool is most widely used in research today, but the one that’s most talked about is undoubtedly Git. This is largely because of GitHub, a popular hosting site that combines the technical infrastructure for collaboration via Git with a modern web interface. GitHub is free for public and open source projects and for users in academia and nonprofits. GitLab is a well-regarded alternative that some prefer, because the GitLab platform itself is free and open source. Bitbucket provides free hosting for both Git and Mercurial repositories, but does not have nearly as many scientific users. CSIRO hosts it’s own instance of BitBucket for employee use.
How Version Control Systems Work
A version control system stores snapshots of a project’s files in a repository. Users modify their working copy of the project, and then save changes to the repository when they wish to make a permanent record and/or share their work with colleagues. The version control system automatically records when the change was made and by whom along with the changes themselves.
Crucially for collaboration, if several people have edited files simultaneously, the version control system will detect the collision and require them to resolve any conflicts before recording the changes. Modern version control systems also allow repositories to be synchronized with each other, so that no one repository becomes a single point of failure. Tool-based version control has several benefits over manual version control:
Instead of requiring users to make backup copies of the whole project, version control safely stores just enough information to allow old versions of files to be re-created on demand.
Instead of relying on users to choose sensible names for backup copies, the version control system timestamps all saved changes automatically.
Instead of requiring users to be disciplined about completing the changelog, version control systems prompt them every time a change is saved. They also keep a 100% accurate record of what was actually changed, as opposed to what the user thought they changed, which can be invaluable when problems crop up later.
Instead of simply copying files to remote storage, version control checks to see whether doing that would overwrite anyone else’s work. If so, they facilitate identifying conflict and merging changes.
Changelog in action
Have a look at one of the example github repositories and how they track changes:
Give examples of:
- what makes their changelogs good?
- what could be improved?
Also, what would be the most difficult feature to replicate with manual version control?
Some good things:
- all log entries contain date and author
- all log entries contain list of files that have been modified
- for text files the actual change can be visible
- the description text gives an idea of the change
Some things that could be improved:
- The pigs files should probably be recorded in smaller chunks (commits). The raw data and cleaned data could be added separetely unless they all were captured at the same time.
- Rather than general “Readme update” a more specific descriptin could be provied “Reformated headers and list”
- Some of the Ballou et al changes could do with more detailed descriptions, for example why the change took place in case of IQ_TREE entries
Something difficult to replicate manually:
- The changelog is linked to a complete description of the file changes.
- Click on an entry, for example
Clarify README.mdorupdate readme file, and you’ll see the file changes with additions marked with + (in green) and deletions marked with - (in red).
What Not to Put Under Version Control
The benefits of version control systems don’t apply equally to all
file types. In particular, version control can be more or less rewarding
depending on file size and format. First, file comparison in version
control systems is optimized for plain text files, such as source code.
The ability to see so-called “diffs” is one of the great joys of version
control systems. Unfortunately, Microsoft Office files (like the
.docx files used by Word) or other binary files, e.g.,
PDFs, can be stored in a version control system, but it is not always
possible to pinpoint specific changes from one version to the next.
Tabular data (such as CSV files) can be put in version control, but
changing the order of the rows or columns will create a big change for
the version control system, even if the data itself has not changed.
Second, raw data should not change, and therefore should not require version tracking. Keeping intermediate data files and other results under version control is also not necessary if you can re-generate them from raw data and software. However, if data and results are small, we still recommend versioning them for ease of access by collaborators and for comparison across versions.
Third, today’s version control systems are not designed to handle megabyte-sized files, never mind gigabytes, so large data or results files should not be included. (As a benchmark for “large”, the limit for an individual file on GitHub is 100MB.) Some emerging hybrid systems such as Git LFS put textual notes under version control, while storing the large data itself in a remote server, but these are not yet mature enough for us to recommend.
Inadvertent Sharing
Researchers dealing with data subject to legal restrictions that prohibit sharing (such as medical data) should be careful not to put data in public version control systems. Some institutions may provide access to private version control systems, so it is worth checking with your IT department.
Additionally, be sure not to unintentionally place security credentials, such as passwords and private keys, in a version control system where it may be accessed by others.
Attribution
This episode was adapted from and includes material from Wilson et al. Good Enough Practices for Scientific Computing.
Key Points
- Small, frequent changes are easier to track
- Tracking change systematically with checklists is helpful
- Version control systems help adhere to good practices
Content from Manuscripts
Last updated on 2024-01-29 | Edit this page
Estimated time: 20 minutes
Overview
Questions
- How do I write a collaborative paper?
Objectives
- Know how to frame writing manuscripts as a computing project
- Compare the benefits and drawbacks of 3 approaches
- Know where to start with text-based version control as a good practice for writing manuscripts
An old joke says that doing the research is the first 90% of any project; writing up is the other 90%. While writing is rarely addressed in discussions of scientific computing, computing has changed scientific writing just as much as it has changed research.
Writing manuscripts is often collaborative, and so a team with diverse backgrounds, skills, and expectations must work together. In our experience, setting explicit expectations for writing is essential, just like other collaborations.
Discussion (3 mins)
Whether or not you have written a scientific manuscript before, you probably have experience of group work or writing. Discuss on the collaborative document:
- What tools have you used before for group writing?
- What’s gone wrong with group writing you’ve been involved with in the past?
The First Rule Is…
The workflow you choose is less important than having all authors agree on the workflow before writing starts. Make sure to also agree on a single method to provide feedback, be it an email thread or mailing list, an issue tracker, or some sort of shared online to-do list.
We suggest having a meeting (or online thread) of all authors at the beginning of the writing process. Ask everyone how they would prefer to write a manuscript. Then agree on a decision and process, and put the outcome in writing. If co-authors are learning new tools, ask someone familiar with those tools to support them!
Making email-based workflows work
A common practice in academic writing is for the lead author to email successive versions of a manuscript to coauthors to collect feedback, which is returned as changes to the document, comments on the document, plain text in email, or a mix of all three. This allows co-authors to use familiar tools, but results in a lot of files to keep track of, and a lot of tedious manual labor to merge comments to create the next master version.
However, if a (senior) co-author insists on using a particular format, like word or LaTeX, or on sending comments by email or written on printouts, in our experience it can be very difficult to convince them to change. Two principles make an email-based workflow work: informative filenames with date and initials, and a single lead author who co-ordinates.
Top tips for writing manuscripts via email
- Give your manuscript file an informative name, and update the date and initials of last edit, for example
best_practices_manuscript_2013-12-01_GW.docwould be the version edited by GW on 1st December 2013.- Choose one person to co-ordinate (i.e. the lead author), who is responsible for merging comments and sending out updated manuscripts to all other co-authors.
Good practices beyond an email-based workflow
Instead of an email-based workflow, we recommend mirroring good practices for managing software and data to make writing scalable, collaborative, and reproducible. As with our recommendations for version control in general, we suggest that groups choose one of two different approaches for managing manuscripts. The goals of both are to:
Ensure that text is accessible to yourself and others now and in the future by making a single master document that is available to all coauthors at all times.
Reduce the chances of work being lost or people overwriting each other’s work.
Make it easy to track and combine contributions from multiple collaborators.
Avoid duplication and manual entry of information, particularly in constructing bibliographies, tables of contents, and lists.
Make it easy to regenerate the final published form (e.g., a PDF) and to tell if it is up to date.
Make it easy to share that final version with collaborators and to submit it to a journal.
Single Master Online
Our first alternative will already be familiar to many researchers:
- Write manuscripts using online tools with rich formatting, change tracking, and reference management, such as Google Docs or MS OneDrive. With the document online, everyone’s changes are in one place, and hence don’t need to be merged manually.
This is easy under our current Microsoft Office organisational setup, where Word documents (and others) may be converted to shared online documents automatically when sharing through Outlook or Teams.
Text-based Documents Under Version Control
The second approach treats papers exactly like software, and has been used by researchers in mathematics, astronomy, physics, and related disciplines for decades:
- Write the manuscript in a plain text format that permits version control such as LaTeX or Markdown, and then convert them to other formats such as PDF as needed using scriptable tools like Pandoc.
Using a version control system provides good support for finding and merging differences resulting from concurrent changes. It also provides a convenient platform for making comments and performing review.
This approach re-uses the version control tools and skills used to manage data and software, and is a good starting point for fully-reproducible research. However, it requires all contributors to understand a much larger set of tools, including markdown or LaTeX, make, BiBTeX, and Git/GitHub.
It is even possible using this approach to combine manuscripts and data analysis, e.g. through Rmarkdown.
Top tips for writing manuscripts via text-based version control
- Project organization is crucial here, structure your folder thoughtfully.
- Make a project/manuscript README file, including the agreed workflow.
- Separate sentences by linebreaks in your plain-text document, to make comparisons and merging easier.
Benefits and drawbacks of each approach
| Things to consider | Email based workflow | Single master online | Text-based under version control |
|---|---|---|---|
| Previous user experience/comfort | High | Medium | Low |
| Visible tracking of changes | Low | Variable | High |
| Institutional support | Low | High | Low |
| Ease of merging changes and suggestions | Low | Medium | High |
| Distributed control | Low | High | High |
| Ease of formatting changes for re-submission | Low | Low | High |
While we feel that text-based version control is a superior method, the barriers to entry may be too high for many users. The single master online approach is a good compromise. If your institution has invested in an environment (Google Docs / MS Office), users can stay within their familiar desktop GUI applications while still taking advantage of automatic file versioning and shared editing.
Discussion: Approaching your next manuscript (7 mins)
In groups, discuss:
- What’s the next manuscript you’ll work on, and who with?
- Which approaches will you use to collaborate on this next manuscript?
Getting started writing text-based version control
Manubot is an open-source system for writing scholarly manuscripts via GitHub, with tutorials.
Supplementary Materials
Supplementary materials often contain much of the work that went into the project, such as tables and figures or more elaborate descriptions of the algorithms, software, methods, and analyses. In order to make these materials as accessible to others as possible, do not rely solely on the PDF format, since extracting data from PDFs is notoriously hard. For the same reason, Excel is not a suitable file format for table data that others may want to re-analyze. It is acceptable for summary statistics tables so long as the underlying data is also available in a text file format such as CSV.
We recommend separating the results that you may expect others to reuse (e.g., data in tables, data behind figures) into separate, text-format files in formats such as CSV, JSON, YAML, XML, or HDF5. The same holds for any commands or code you want to include as supplementary material: use the format that most easily enables reuse (source code files, Unix shell scripts etc).
Attribution
This episode was adapted from and includes material from Wilson et al. Good Enough Practices for Scientific Computing.
Key Points
- Have all authors agree on a workflow before the writing starts
- Email-based workflows work better with informative filenames and clear co-ordination
- Text-based documents with version control scale better, if co-authors are familiar with the tools
- Single Master Online approaches can be an effective compromise
Content from What To Do Next
Last updated on 2023-08-31 | Edit this page
Estimated time: 10 minutes
Overview
Questions
- How can you improve practices after this workshop?
Objectives
- Learn about other resources to improve good practices
- Reflect on good enough practices appropriate for your career stage
Heads up
When you lose your data, or stare, disgusted, at a horrible file, think of today.
Learning good practices is a long-term process
We have outlined a series of good enough practices for scientific computing. These practices are pragmatic, broadly accessible, and can be applied by both individuals and groups. Most importantly, these practices make researchers more productive individually by enabling them to get more done in less time and with less pain. They also accelerate research as a whole by making work more reproducible.
We have covered good practices in several areas:
- Data management
- Software
- Collaboration
- Project organization
- Keeping track of changes
- Manuscripts
We have explained the shared set of principles behind good practices:
- Planning
- Modular organization
- Names
- Documentation
Collectively, these practices make your work easier to use for other people, including for your future self. Thinking through your work from a collaborator’s point of view is helpful, and ultimately kinder to your future self.
What we left out, and where to learn more
Learning good practices is a long-term process that never stops. We left out many good practices that, although useful, have more niche applications.
We recommend the paper Best Practices in Scientific Computing, especially for those gaining more experience with coding. There are many other useful papers and resources that we have selected.
One of your best resources is your colleagues - talk to them! In our experience, people who have learned good practices themselves are eager to teach them to others.
What other resources are available?
- Where can you look for more help?
- What resources are available in your research group? Your institution?
Different people make different contributions to good practice
Progress in computational good practices comes from different places in the scientific community:
- Funders can require data management plans and provide support for data management.
- Institutions can value the time that goes into data sharing and training.
- Research librarians, research software engineers, and data managers support others in learning and applying good practices.
- PIs and lab heads can require that lab members share code and data, and make it easy for them to do so.
- Lab members can organise “data curation days” and training sessions to share good practices.
- Self-organised groups led by students and postdocs can share ideas and train each other.
- Global organisations like The Carpentries can co-ordinate training and support training materials.
- Professional societies can help to organise training.
It takes time to learn good practices, and time to train others. This time has to be valued by individuals, funders, institutions, and the scientific community.
Developing your own good practices is ultimately a personal journey that will impact your career and impact the people around you. Maybe today you will learn to write README files, or next year you might start using version control, or in the future you could find yourself teaching students to write data management plans for their projects.
What will you do next?
- Which good enough practices will you use in your next project?
- How can you help your colleagues in using good practices?
- What could your longer-term contribution to good practices be?
Attribution
Content of this episode was adopted after Wilson et al. Good Enough Practices for Scientific Computing.
Key Points
- Learning good practices is a long-term process
- Different people make different contributions to good practice
Content from Agile
Last updated on 2024-01-31 | Edit this page
Estimated time: 60 minutes
What is ‘Agile’
‘Agile’ is a project management methodology, particularly for software development, built around a 4 point philosophical manifesto and a 12 point set of principles.
Agile is typified by small teams that self-organise (‘scrum’) on how they will address a backlog of requested work, in short cycles (‘sprints’), by breaking problems into small tasks, with frequent feedback and result delivery. It is a highly iterative approach to planning, that allows for high flexibility and less forward planning. A sprint may last 1-4 weeks, in which time an entire cycle of planning, designing, implementing, testing and delivering takes place, with small tasks hopefully addressed to completion, followed by a review and retrospective that may or may not end up influencing the next sprint cycle.
Atlassian on scrums, Kanban and Jira visualisations
Key Points
- The Agile approach is to break problems into smaller tasks and fully address them in short, iterative work cycles (sprints), with each cycle ending in review and discussion before planning the next cycle.
- Aspects of this approach may be useful in data science work.
